Neural Networks using R
Using neuralnet
neuralnet depends on two other packages: grid and MASS (Venables and Ripley, 2002). It is used is primarily with functions dealing with regression analyses like linear models (lm) and general linear models (glm). As essential arguments, we must specify a formula in terms of response variables ~ sum of covariates and a data set containing covariates and response variables. Default values are defined for all other parameters (see next subsection). We use the data set infert that is provided by the package datasets to illustrate its application. This data set contains data of a case-control study that investigated infertility after spontaneous and induced abortion (Trichopoulos et al., 1976). The data set consists of 248 observations, 83 women, who were infertile (cases), and 165 women, who were not infertile (controls). It includes amongst others the variables age, parity, induced, and spontaneous. The variables induced and spontaneous denote the number of prior induced and spontaneous abortions, respectively. Both variables take possible values 0, 1, and 2 relating to 0, 1, and 2 or more prior abortions. The age in years is given by the variable age and the number of births by parity.Training of neural networks
The function neuralnet, used for training a neural network, provides the opportunity to define the required number of hidden layers and hidden neurons according to the needed complexity. The complexity of the calculated function increases with the addition of hidden layers or hidden neurons. The default value is one hidden layer with one hidden neuron. The most important arguments of the function are the following:- formula, a symbolic description of the model to be fitted (see above). No default.
- data, a data frame containing the variables specified in formula. No default.
- hidden, a vector specifying the number of hidden layers and hidden neurons in each layer. For example the vector (3,2,1) induces a neural network with three hidden layers, the first
- startweights, a vector containing prespecified starting values for the weights. Default: random numbers drawn from the standard normal distribution
- algorithm, a string containing the algorithm type. Possible values are “backprop“, “rprop+”, “rprop-“, “sag“, or “slr“. “backprop” refers to traditional backpropagation, “rprop+” and “rprop-” refer to resilient backpropagation with and without weight backtracking and “sag” and “slr” refer to the modified globally convergent algorithm (gr-prop). “sag” and “slr” define the learning rate that is changed according to all others. “sag” refers to the smallest absolute derivative, “slr” to the smallest learning rate. Default: “rprop+”
- err.fct, a differentiable error function. The strings “sse” and “ce” can be used, which refer to ‘sum of squared errors’ and ‘cross entropy’. Default: “sse“
- act.fct, a differentiable activation function. The strings “logistic” and “tanh” are possible for the logistic function and tangens hyperbolicus (that is the Latin name for hyperbolic tangent). Default: “logistic“
- linear.output, logical. If act.fct should not be applied to the output neurons, linear.output has to be TRUE. Default: TRUE
- likelihood, logical. If the error function is equal to the negative log-likelihood function, likelihood has to be TRUE. Akaike’s Information Criterion (AIC, Akaike, 1973) and Bayes Information Criterion (BIC, Schwarz, 1978) will then be calculated. Default: FALSE
- exclude, a vector or matrix specifying weights that should be excluded from training. A matrix with n rows and three columns will exclude n weights, where the first column indicates the layer, the second column the input neuron of the weight, and the third neuron the output neuron of the weight. If given as vector, the exact numbering has to be known. The numbering can be checked using the provided plot or the saved starting weights. Default: NULL
- constant.weights, a vector specifying the values of weights that are excluded from training and treated as fixed. Default: NULL
> library(neuralnet)
Loading required package: grid
Loading required package: MASS
> nn nn
Call:
neuralnet(formula = case~age+parity+induced+spontaneous, data = infert, hidden = 2, err.fct = "ce", linear.output = FALSE)
1 repetition was calculated.
Error Reached Threshold Steps1 125.2126851 0.008779243419 5254
We save basic information about the training process and the trained neural network in nn. This includes all information that has to be known to reproduce the results as for instance the starting weights. Important values are the following:- net.result, a list containing the overall result, i.e. the output, of the neural network for each replication.
- weights, a list containing the fitted weights of the neural network for each replication.
- generalized.weights, a list containing the generalized weights of the neural network for each replication.
- result.matrix, a matrix containing the error, reached threshold, needed steps, AIC and BIC (computed if likelihood=TRUE) and estimated weights for each replication. Each column represents one replication.
- startweights, a list containing the starting weights for each replication.
> nn$result.matrix
1 error 125.212685099732
reached.threshold 0.008779243419
steps 5254.000000000000
Intercept.to.1layhid1 5.593787533788
age.to.1layhid1 -0.117576380283
parity.to.1layhid1 1.765945780047
induced.to.1layhid1 -2.200113693672
spontaneous.to.1layhid1 -3.369491912508
Intercept.to.1layhid2 1.060701883258
age.to.1layhid2 2.925601414213
parity.to.1layhid2 0.259809664488
induced.to.1layhid2 -0.120043540527
spontaneous.to.1layhid2 -0.033475146593
Intercept.to.case 0.722297491596
1layhid.1.to.case -5.141324077052
1layhid.2.to.case 2.623245311046
The training process needed 5254 steps until all absolute partial derivatives of the error function were smaller than 0.01 (the default threshold). The estimated weights range from −5.14 to 5.59. For instance, the intercepts of the first hidden layer are 5.59 and 1.06 and the four weights leading to the first hidden neuron are estimated as −0.12, 1.77, −2.20, and −3.37 for the covariates age, parity, induced and spontaneous, respectively. If the error function is equal to the negative log-likelihood function, the error refers to the likelihood as is used for example to calculate Akaike’s Information Criterion (AIC).We save the given data in nn$covariate and nn$response as well as in nn$data for the whole data set inclusive non-used variables. The output of the neural network, i.e. the fitted values o(x), is provided by nn$net.result:
> out
> dimnames(out) head(out)
Age parity induced spontaneous nn-output
[1,] 26 6 1 2 0.1519579877
[[2,] 42 1 1 0 0.6204480608
[[3,] 39 6 2 0 0.1428325816
[[4,] 34 4 2 0 0.1513351888
[[5,] 35 3 1 1 0.3516163154
[[6,] 36 4 2 1 0.4904344475
In this case, the object nn$net.result is a list consisting of only one element relating to one calculated replication. If more than one replication were calculated, we would save the each of the outputs in a separate list element. This approach is the same for all values that change with replication apart from net.result that is saved as matrix with one column for each replication.To compare the results, neural networks are trained with the same parameter setting as above using neuralnet with algorithm=”backprop” and the package nnet.
nn.bp and nn.nnet show equal results. Both training processes last only a very few iteration steps and the error is approximately 158. Thus, in this little comparison, the model fit is less satisfying than that achieved by resilient backpropagation.> nn.bp nn.bp
Call:
neuralnet(formula = case~age+parity+induced+spontaneous, data = infert, hidden = 2, learningrate = 0.01, algorithm = "backprop", err.fct = "ce", linear.output = FALSE)
1 repetition was calculated.
Error Reached Threshold Steps
1 158.085556 0.008087314995 4
> nn.nnet # weights: 13
initial value 256.848809
iter 10 value 158.085559
final value 158.085463
converged
neuralnet includes the calculation of generalized weights as introduced by Intrator and Intrator (2001). The generalized weight w̃i is defined as the contribution of the ith covariate to the log-odds:
The generalized weight expresses the effect of each covariate xi and thus has an analogous interpretation as the ith regression parameter in regression models. However, the generalized weight depends on all other covariates. Its distribution indicates whether the effect of the covariate is linear since a small variance suggests a linear effect (Intrator and Intrator, 2001). They are saved in nn$generalized.weights and are given in the following format (rounded values)
The columns refer to the four covariates age (j =1), parity (j = 2), induced (j = 3), and spontaneous(j = 4) and a generalized weight is given for each observation even though they are equal for each covariate combination.> head(nn$generalized.weights[[1]])
[,1] [,2] [,3] [,4]
1 0.0088556 -0.1330079 0.1657087 0.2537842
2 0.1492874 -2.2422321 2.7934978 4.2782645
3 0.0004489 -0.0067430 0.0084008 0.0128660
4 0.0083028 -0.1247051 0.1553646 0.2379421
5 0.1071413 -1.6092161 2.0048511 3.0704457
6 0.1360035 -2.0427123 2.5449249 3.8975730
Visualizing the results
The results of the training process can be visualized by two different plots. First, the trained neural network can simply be plotted by
> plot(nn)
The resulting plot is given in Figure 1.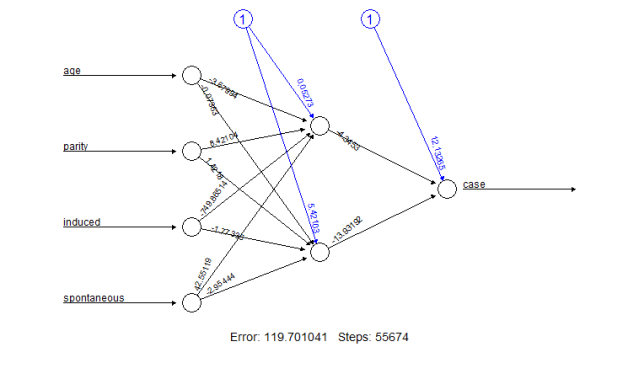
Figure 1: Plot of a trained neural network including trained synaptic weights and basic information about the training process.
It reflects the structure of the trained neural network, i.e. the network topology. The plot includes by default the trained synaptic weights, all intercepts as well as basic information about the training process like the overall error and the number of steps needed to converge. Especially for larger neural networks, the size of the plot and that of each neuron can be determined using the parameters dimension and radius, respectively.
The second possibility to visualize the results is to plot generalized weights. gwplot uses the calculated generalized weights provided by nn$generalized.weights and can be used by the following statements:
> par(mfrow=c(2,2))
> gwplot(nn, selected.covariate="age", min=-2.5, max=5)
> gwplot(nn, selected.covariate="parity",
+ min=-2.5, max=5)
> gwplot(nn, selected.covariate="induced",
+ min=-2.5, max=5)
> gwplot(nn, selected.covariate="spontaneous",
+ min=-2.5, max=5)
The corresponding plot is shown in Figure 2.
Figure 2: Plots of generalized weights with respect to each covariate.
The generalized weights are given for all covariates within the same range. The distribution of the generalized weights suggests that the covariate age has no effect on the case-control status since all generalized weights are nearly zero and that at least the two covariates induced and spontaneous have a non-linear effect since the variance of their generalized weights is overall greater than one.
Additional features
The compute function
compute calculates and summarizes the output of each neuron, i.e. all neurons in the input, hidden and output layer. Thus, it can be used to trace all signals passing the neural network for given covariate combinations. This helps to interpret the network topology of a trained neural network. It can also easily be used to calculate predictions for new covariate combinations. A neural network is trained with a training data set consisting of known input-output pairs. It learns an approximation of the relationship between inputs and outputs and can then be used to predict outputs o(xnew ) relating to new covariate combinations xnew . The function compute simplifies this calculation. It automatically redefines the structure of the given neural network and calculates the output for arbitrary covariate combinations.To stay with the example, predicted outputs can be calculated for instance for missing combinations with age=22, parity=1, induced ≤ 1, and spontaneous ≤ 1. They are provided by new.output$net.result
This means that the predicted probability of being a case given the mentioned covariate combinations, i.e. o(x), is increasing in this example with the number of prior abortions.> new.output new.output$net.result
[,1]
[1,] 0.1477097
[2,] 0.1929026
[3,] 0.3139651
[4,] 0.8516760
The confidence.interval function
The weights of a neural network follow a multivariate normal distribution if the network is identified (White, 1989). A neural network is identified if it does not include irrelevant neurons, neither in the input layer nor in the hidden layers. An irrelevant neuron in the input layer can be, for instance, a covariate that has no effect or that is a linear combination of other included covariates. If this restriction is fulfilled and if the error function equals the negative log-likelihood, a confidence interval can be calculated for each weight. The neuralnet package provides a function to calculate these confidence intervals regardless of whether all restrictions are fulfilled. Therefore, we have to be careful interpreting the results.Since the covariate age has no effect on the outcome and the related neuron is thus irrelevant, a new neural network (nn.new), which has only the three input variables parity, induced, and spontaneous, has to be trained to demonstrate the usage of confidence.interval. Let us assume that all restrictions are now fulfilled, i.e. neither the three input variables nor the two hidden neurons are irrelevant. Confidence intervals can then be calculated with the function confidence.interval:
> nn.new nn.new
Call: neuralnet(formula = case ~ parity + induced + spontaneous, data = infert, hidden = 2, err.fct = "ce", linear.output = FALSE)
1 repetition was calculated.
Error Reached Threshold Steps
1 123.7940021 0.009743898478 2774
> nn.new$result.matrix1error 123.794002097050
reached.threshold 0.009743898478
steps 2774.000000000000
Intercept.to.1layhid1 -9.024035981211
parity.to.1layhid1 -9.355649895691
induced.to.1layhid1 4.604728544071
spontaneous.to.1layhid1 21.693583835740
Intercept.to.1layhid2 -7.982974207707
parity.to.1layhid2 -3.335661191123
induced.to.1layhid2 14.149057774749
spontaneous.to.1layhid2 7.372698725819
Intercept.to.case -2.227015901571
1layhid.1.to.case 2.805901061643
1layhid.2.to.case 1.769454351740> out dimnames(out) head(out)
parity induced spontaneous nn.new-output
[1,] 6 1 2 0.2748949333
[2,] 1 1 0 0.3644454394
[3,] 6 2 0 0.2296907484
[4,] 4 2 0 0.3871712281
[5,] 3 1 1 0.3757109614
[6,] 4 2 1 0.3875643222
> head(nn.new$generalized.weights[[1]])
[,1] [,2] [,3]
1 -1.214196782891496 5.15032191028160 2.68369804196508
2 -0.310485049736654 1.31689903088964 0.68625753443013
3 -1.442613638423590 6.11921371721348 3.18855996591000
4 -0.005521928145309 0.02342266671564 0.01220493037762
5 -0.163520412577241 0.69159093349938 0.36148214704463
6 -0.000008138035933 0.00001703598866 0.00001849315091
> plot(nn.new)
> new.output new.output$net.result
[,1]
[1,] 0.09735243992
[2,] 0.36444543943
[3,] 0.91271565323
> ci ci$lower.ci[[1]]
[[1]][[1]]
[,1] [,2]
[1,] -18.323535695 -18.920799122
[2,] -17.278014756 -9.526947243
[3,] 1.659472408 -9.344351919
[4,] 2.390600705 -4.210395184[[1]][[2]]
[,1]
[1,] -2.9311558742
[2,] 1.8202683312
[3,] 0.9017336937
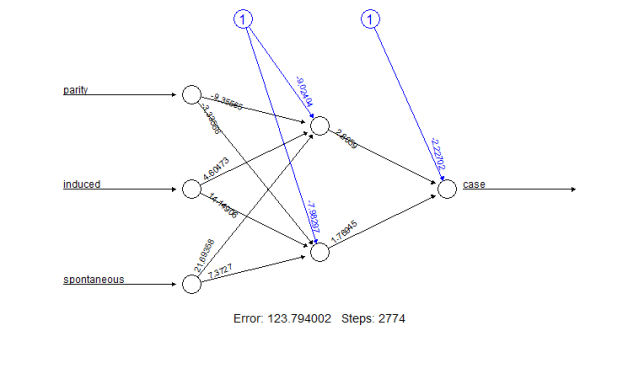
Figure 3: Plot of the new trained neural network including trained synaptic weights and basic information about the training process.
For each weight, ci$lower.ci provides the related lower confidence limit and ci$upper.ci the related upper confidence limit. The first matrix contains the limits of the weights leading to the hidden neurons. The columns refer to the two hidden neurons. The other three values are the limits of the weights leading to the output neuron.
Summary
This paper gave a brief introduction to multilayer perceptrons and supervised learning algorithms. It introduced the package neuralnet that can be applied when modeling functional relationships between covariates and response variables. neuralnet contains a very flexible function that trains multilayer perceptrons to a given data set in the context of regression analyses. It is a very flexible package since most parameters can be easily adapted. For example, the activation function and the error function can be arbitrarily chosen and can be defined by the usual definition of functions in R.Bibliography
- Akaike. Information theory and an extension of the maximum likelihood principle. In Petrov BN and Csaki BF, editors, Second international symposium on information theory, pages 267–281. Academiai Kiado, Budapest, 1973.
- Almeida, C. Baugh, C. Lacey, C. Frenk, G. Granato, L. Silva, and A. Bressan. Modeling the dsty universe i: Introducing the artificial neural network and first applications to luminosity and color distributions. Monthly Notices of the Royal Astronomical Society, 402:544–564, 2010.
- Anastasiadis, G. Magoulas, and M. Vrahatis. New globally convergent training scheme based on the resilient propagation algorithm. Neurocomputing, 64:253–270, 2005.
- Bishop. Neural networks for pattern recognition. Oxford University Press, New York, 1995.
- Fritsch and F. Günther. neuralnet: Training of Neural Networks. R Foundation for Statistical Computing, 2008. R package version 1.2.
- Günther, N. Wawro, and K. Bammann. Neural networks for modeling gene-gene interactions in association studies. BMC Genetics, 10:87, 2009. http://www.biomedcentral.com/1471-2156/10/87.
- Hornik, M. Stichcombe, and H. White. Multilayer feedforward networks are universal approximators. Neural Networks, 2:359–366, 1989.
- Intrator and N. Intrator. Interpreting neural-network results: a simulation study. Computational Statistics & Data Analysis, 37:373–393, 2001.
- Kumar and D. Zhang. Personal recognition using hand shape and texture. IEEE Transactions on Image Processing, 15:2454–2461, 2006.
- C. Limas, E. P. V. G. Joaquín B. Ordieres Meré, F. J. M. de Pisón Ascacibar, A. V. P. Espinoza, and F. A. Elías. AMORE: A MORE Flexible Neural Network Package, 2007. URL http://wiki.r-project. org/rwiki/doku.php?id=packages:cran:amore. R package version 0.2-11.
- McCullagh and J. Nelder. Generalized Linear Models. Chapman and Hall, London, 1983.
- Riedmiller. Advanced supervised learning in multilayer perceptrons – from backpropagation to adaptive learning algorithms. International Journal of Computer Standards and Interfaces, 16:265–278,1994.
- Riedmiller and H. Braun. A direct method for faster backpropagation learning: the rprop algorithm. Proceedings of the IEEE International Conference on Neural Networks (ICNN), 1:586–591, 1993.
- Rocha, P. Cortez, and J. Neves. Evolutionary neu- ral network learning. Lecture Notes in Computer Science, 2902:24–28, 2003.
- Rojas. Neural Networks. Springer-Verlag, Berlin, 1996.
- Schiffmann, M. Joost, and R. Werner. Optimization of the backpropagation algorithm for training multilayer perceptrons. Technical report, University of Koblenz, Insitute of Physics, 1994.
- Schwarz. Estimating the dimension of a model. Ann Stat, 6:461–464, 1978.
- Trichopoulos, N. Handanos, J. Danezis, A. Kalandidi, and V. Kalapothaki. Induced abortion and secondary infertility. British Journal of Obstetrics and Gynaecology, 83:645–650, 1976.
- Venables and B. Ripley. Modern Applied Statistics with S. Springer, New York, fourth edition, 2002. URL http://www.stats.ox.ac.uk/ pub/MASS4. ISBN 0-387-95457-0.
- White. Learning in artificial neural networks: a statistical perspective. Neural Computation, 1:425–464, 1989.

Authored by:Jeffrey Strickland, Ph.D.
Jeffrey Strickland, Ph.D., is the Author of Predictive Analytics Using R and a Senior Analytics Scientist with Clarity Solution Group. He has performed predictive modeling, simulation and analysis for the Department of Defense, NASA, the Missile Defense Agency, and the Financial and Insurance Industries for over 20 years. Jeff is a Certified Modeling and Simulation professional (CMSP) and an Associate Systems Engineering Professional (ASEP). He has published nearly 200 blogs on LinkedIn, is also a frequently invited guest speaker and the author of 20 books including:
- Operations Research using Open-Source Tools
- Discrete Event simulation using ExtendSim
- Crime Analysis and Mapping
- Missile Flight Simulation
- Mathematical Modeling of Warfare and Combat Phenomenon
- Predictive Modeling and Analytics
- Using Math to Defeat the Enemy
- Verification and Validation for Modeling and Simulation
- Simulation Conceptual Modeling
- System Engineering Process and Practices


Great post thanks for sharing
ReplyDeleteazure certification training in chennai
Neural networks are a powerful tool for various machine learning tasks, such as classification, regression, and pattern recognition. In R, there are several packages available to build and train neural networks. Some of the most commonly used packages include nnet, neuralnet, and keras.
DeleteHere’s a basic guide to getting started with neural networks in R using these packages:
1. Using the nnet Package
The nnet package is part of the base R distribution and is suitable for small to moderately sized datasets.
Deep Learning Projects for Final Year
Machine Learning Projects for Final Year
Artificial Intelligence Projects For Final Year
Great blog!!! This is a very different and unique content. I am waiting for your another post...
ReplyDeleteSocial Media Marketing Courses in Chennai
Social Media Marketing Training in Chennai
Embedded System Course Chennai
Linux Training in Chennai
Tableau Training in Chennai
Spark Training in Chennai
Oracle Training in Chennai
Oracle DBA Training in Chennai
Social Media Marketing Courses in Chennai
Social Media Marketing Training in Chennai
perde modelleri
ReplyDeletesms onay
TURKCELL MOBİL ÖDEME BOZDURMA
Nft nasıl alınır
ankara evden eve nakliyat
trafik sigortası
dedektör
web sitesi kurma
aşk kitapları
pendik lg klima servisi
ReplyDeleteümraniye toshiba klima servisi
pendik alarko carrier klima servisi
pendik daikin klima servisi
tuzla toshiba klima servisi
beykoz samsung klima servisi
üsküdar samsung klima servisi
beykoz mitsubishi klima servisi
üsküdar mitsubishi klima servisi
The blog post titled "Neural Networks Using R" by Humalytica provides an introduction to neural networks and how to use them in R. The author explains the basic concepts of neural networks, such as layers, neurons, and activation functions, and walks readers through the process of building a simple neural network in R. The post includes code snippets and visualizations to help readers understand each step of the process. The comments section provides additional insights and tips from readers who have experience working with neural networks in R. Right Brain Training or Flash cards Overall, the blog post offers a helpful starting point for anyone interested in exploring neural networks in R.
ReplyDeleteWhile this article provides a detailed overview of implementing artificial neural networks (ANN) using R, it primarily focuses on the technical aspects of utilizing the neuralnet package for regression analysis. The author's personal preference for using ANN in ensemble models with logistic regression is noted, offering a practical perspective on integrating ANN into predictive modeling workflows. The inclusion of essential arguments and functions of the neuralnet package, along with a demonstration using a case-control study dataset, enhances the article's educational value. Moreover, the article effectively communicates key parameters and considerations for training neural networks, such as the number of hidden layers and neurons, activation functions, and error functions. For more details, visit Right Brain education
ReplyDeleteبهینه سازی یک وب سایت برای سئو انگلیسی شامل روش های استراتژیک برای افزایش موثر ترافیک است. اول از همه، ترکیب کلمات کلیدی مرتبط در سراسر وب سایت ضروری است. با انجام تحقیقات دقیق کلمات کلیدی، می توانید عباراتی را که مخاطبان هدف شما جستجو می کنند شناسایی کنید و به طور طبیعی آنها را در محتوا، متا تگ ها و هدرهای خود ادغام کنید. محتوای جذاب و با کیفیت بالا که در عین رعایت اصول سئو، ارزشی را برای کاربران به ارمغان می آورد، کلید بهبود رتبه بندی موتورهای جستجو است.
ReplyDelete